A series of press releases, including one by Science Publishing, recently announced the first findings of the Avian Phylogenomics Consortium, who analyzed genome sequences and annotation data for 48 bird genomes representing all of the bird taxonomic orders. All of the sequenced genomes, along with any annotation provided by the submitter, are available in NCBI resources including Assembly, Nucleotide, Protein, the Sequence Read Archive (SRA), and BLAST, or from species-specific GenBank genomes FTP directories. RNA-Seq data for some of the bird species can be found in SRA.
With the exception of three very fragmented assemblies, NCBI annotated the genome assemblies submitted by the Avian Phylogenomics Consortium using NCBI’s Eukaryotic Genome Annotation Pipeline, and these annotations are now part of the RefSeq project. The RefSeq project also generated annotations for an additional 6 bird assemblies, for a total of 51 RefSeq genomes. A summary of all the bird genomes that have RefSeq annotation is here.

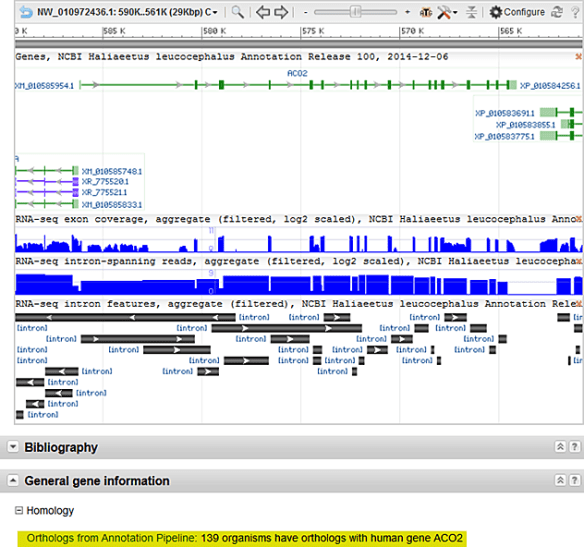
RNA-Seq data was used to generate annotations for 12 of the 51 bird assemblies. The number of protein-coding genes per genome ranges from >13,300 to >21,100 (chicken) with an average of 14,932 protein-coding genes. Orthology to human proteins was also calculated using simple metrics of local synteny and sequence similarity, and on average, roughly 11,000 orthologous proteins were identified per avian genome. These results are shown in the Homology section of NCBI Gene records (see Figure 2 below).
Related news stories:
- Revised Genomes FTP site: More information about GenBank and RefSeq sequence and annotation data on the FTP site.