Join us on June 30, 2021 at 12PM eastern time to learn how to use the new NCBI Datasets resource to find and download gene, genome and SARS-CoV-2 sequence and annotation. You will learn how to access these datasets through either the web interface or the new command-line tools that allow you to incorporate these data in your bioinformatic workflows.
- Date and time: Wed, June 30, 2021 12:00 PM – 12:45 PM EDT
- Register
After registering, you will receive a confirmation email with information about attending the webinar. A few days after the live presentation, you can view the recording on the NCBI webinars playlist on the NLM YouTube channel. You can learn about future webinars on the Webinars and Courses page.

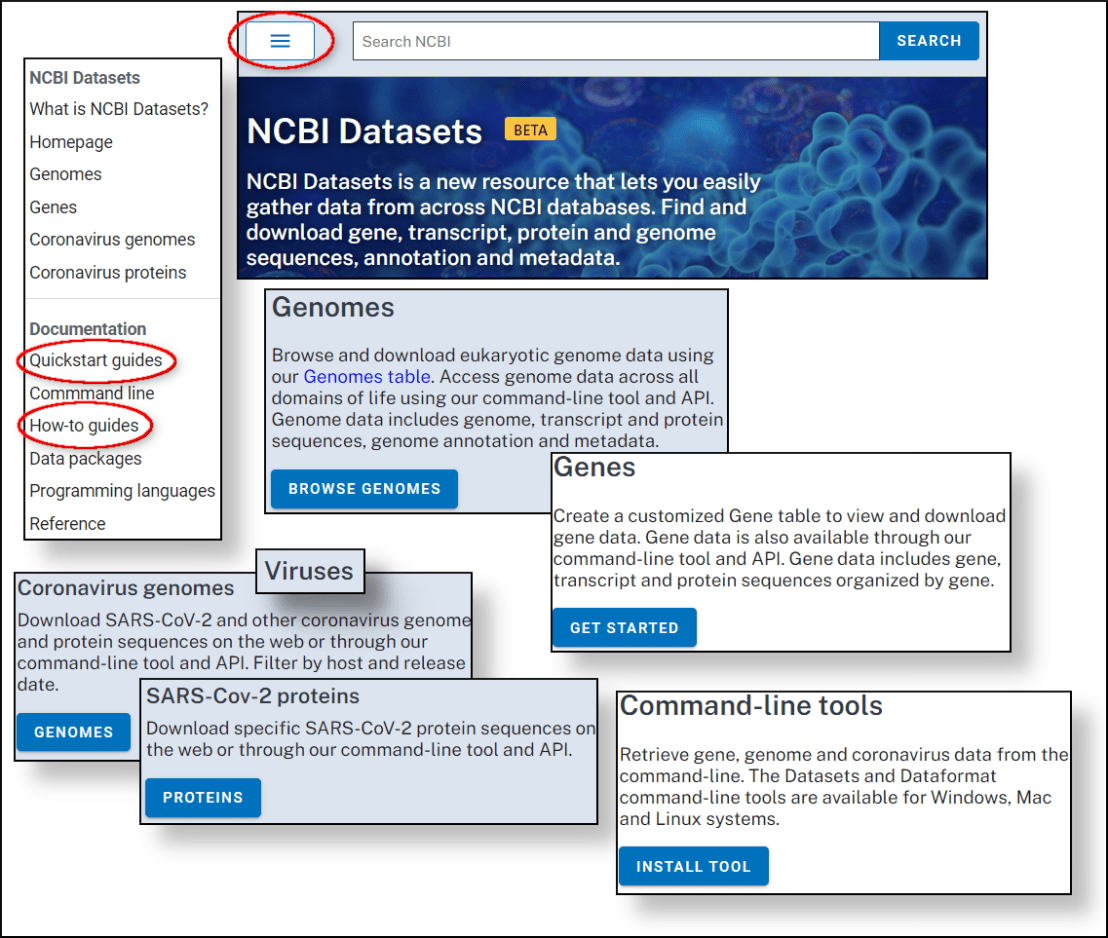

 Figure 1. The
Figure 1. The  Figure 1. The Data tables web download. Top panel. Enter or upload a list of gene identifiers or symbols. Bottom panel. The resulting table display allows you to browse results, download the table or the sequence data for the genes (genomic, transcripts, proteins).
Figure 1. The Data tables web download. Top panel. Enter or upload a list of gene identifiers or symbols. Bottom panel. The resulting table display allows you to browse results, download the table or the sequence data for the genes (genomic, transcripts, proteins). 
 Figure 1. The Assembly page for the Xenopus tropicalis UCB Xtro 10.0 (
Figure 1. The Assembly page for the Xenopus tropicalis UCB Xtro 10.0 (